搜索


Recently, the research team led by Fan Chunhai, Song Ping, and Zuo Xiaolei at Shanghai Jiao Tong University published their latest study titled “Massively parallel homogeneous amplification of chip-scale DNA for DNA information storage (MPHAC-DIS)” in Nature Communications. This research developed a thermodynamically driven high-throughput primer design method (MPHAC), enabling parallel and homogeneous amplification of low-concentration nucleic acid sequences. The DNA storage system built using this method is compatible with multimodal file types including text, images, and videos, enabling random access to any file subset with 99% decoding accuracy.
With global data volumes growing exponentially, DNA has emerged as a critical breakthrough for addressing massive-scale data storage demands due to its ultra-high storage density, long half-life, and low energy consumption. However, current DNA information storage still faces significant challenges, including high costs for DNA synthesis and sequencing. Chip-based large-scale DNA synthesis technology can synthesize millions of DNA molecules simultaneously, significantly reducing synthesis costs. However, its product concentration typically reaches only the femtomolar level, far below the nanomolar concentration required for second-generation sequencing. Traditional amplification methods, due to efficiency variations, are prone to amplification bias, affecting data uniformity and sequencing depth, leading to reduced read accuracy and increased sequencing costs.
To address these challenges, the team developed a thermodynamically controlled uniform amplification method (MPHAC). This approach achieves highly sensitive and uniform amplification of target sequences through stringent thermodynamic energy control, supplemented by GC content screening, secondary structure checks, primer dimer filtering, and BLAST analysis (Figure 1). Compared to traditional fixed-length primer design, simulation results indicate this method exhibits more uniform amplification efficiency (Fold-80 reduced from 3.2 to 1.0) and can reduce costs by up to four orders of magnitude during large-scale random access. The DNA storage system built using this method is compatible with multiple file types and achieves over 99% decoding accuracy. Furthermore, by integrating adversarial neural networks for data repair, it can achieve at least 80% accuracy even at 1× sequencing depth.
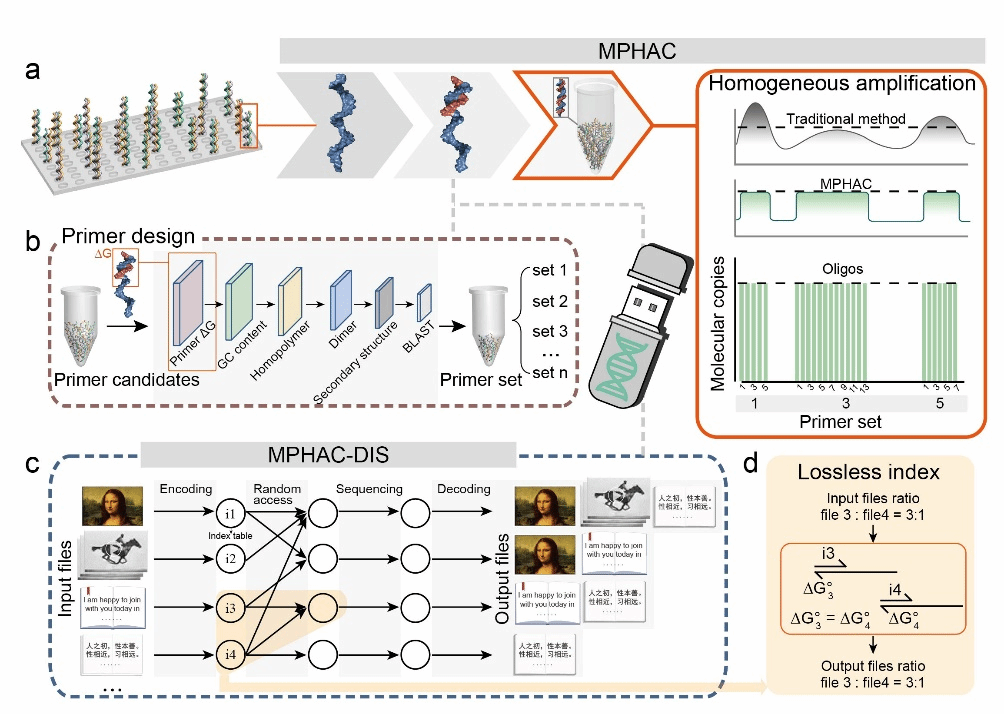
Figure 1 Overview of MPHAC and MPHAC-DIS
The team employed an encoded self-correcting lookup table to encode each byte in ASCII files into a 5-base nucleic acid sequence. By synthesizing 35,406 DNA sequences, they encoded text, images, and video files—including the Three Character Classic, Jiaotong University's Temple Gate, the Mona Lisa, and the moon landing—enabling efficient access and decoding (Figure 2). Leveraging MPHAC's exceptional amplification uniformity and parallelism, this approach holds promise for advancing DNA data storage toward new frontiers in file storage capacity and access throughput.
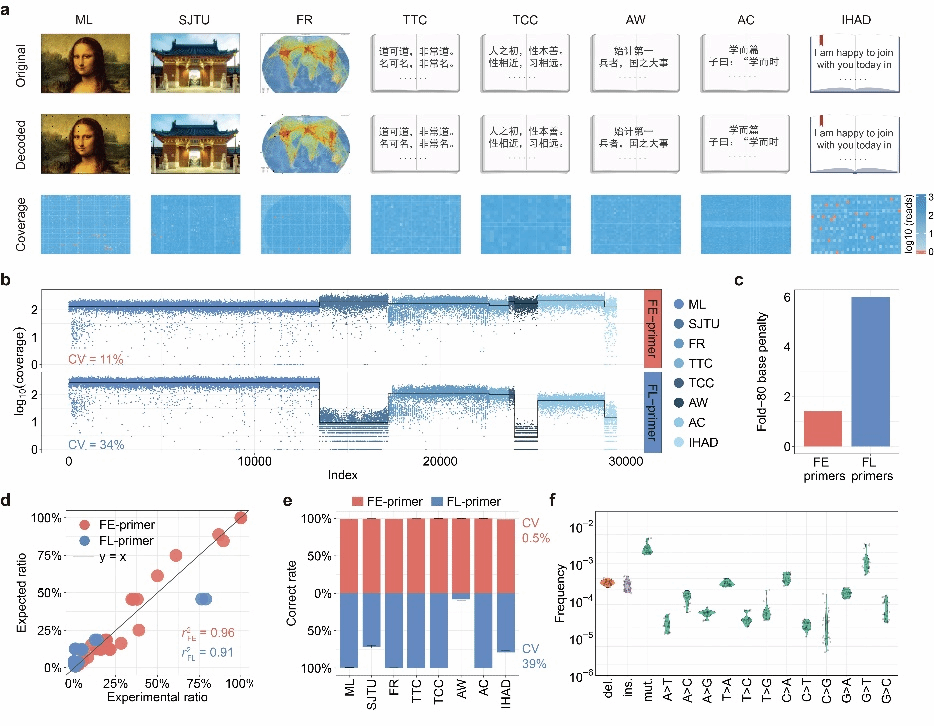
Figure 2 Uniform Amplification and Lossless Indexing of MPHAC-DIS
Weng Zhi, a doctoral candidate at the School of Biomedical Engineering, Shanghai Jiao Tong University, and postdoctoral researcher Li Jiangxue are the first authors of this paper. Corresponding authors include Academician Fan Chunhai from the DNA Storage Research Center, School of Chemistry and Chemical Engineering/Zhangjiang Advanced Research Institute; Associate Professor Song Ping from the DNA Storage Research Center, School of Biomedical Engineering/Zhangjiang Advanced Research Institute; and Professor Zuo Xiaolei from the Molecular Research Institute, Renji Hospital/DNA Storage Research Center, Zhangjiang Advanced Research Institute.
This work was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the Shanghai Municipal Education Commission's “Young Leading Talent Cultivation Program,” and the Shanghai Municipal Science and Technology Innovation Action Plan.
Author: Song Ping Research Group
Contributing Unit: DNA Storage Research Center






 Address:No.1308 Keyuan Road, Pudong District, Shanghai
Address:No.1308 Keyuan Road, Pudong District, Shanghai Phone:86-21-54740000
Phone:86-21-54740000 E-mail:zias@sjtu.edu.cn
E-mail:zias@sjtu.edu.cn