搜索


Recently, the internationally renowned journal Proceedings of the National Academy of Sciences published online collaborative research findings from the Xu Dong Qu Laboratory at the School of Life Sciences and Technology and the Zhangjiang Advanced Research Institute of our university, in partnership with the Zhengyu Zhang Laboratory at Wuhan University and the Binju Wang Laboratory at Xiamen University. The study, titled ‘A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C‒C and C‒N bonds’, was featured in the journal. The paper's co-first authors are Dr Wei Guangzheng (postdoctoral researcher, School of Life Sciences), Mr Duan Borui (PhD candidate, Wuhan University), and Mr Zhou Taiping (PhD candidate, Xiamen University). Corresponding authors include Professor Qu Xudong, Professor Zhang Zhengyu, and Professor Wang Binju.
Cytochrome P450 enzymes, hailed as ‘universal biological catalysts,’ play pivotal roles in drug metabolism, synthesis, and biomanufacturing. Traditionally, P450 enzymes require oxygen, converting it into the active species Compound I (CpdI), which then catalyse oxygenation reactions (e.g., hydroxylation, epoxidation) or radical-driven C-C/C-X bond formation and cleavage reactions (e.g., coupling, cleavage, and rearrangement). However, P450 enzymes often require costly cofactors (such as NADPH) and complex electron-transfer accessory proteins to activate oxygen, limiting their practical applications. Certain P450 enzymes have garnered attention for utilising inexpensive H₂O₂ to form CpdI. However, most can only catalyse simple oxygenation reactions (termed P450 peroxygenases), whilst P450 peroxidases—which catalyse single-electron oxidation to generate radicals without oxygenation—remain relatively rare. This class of activity typically catalyses the formation of C-C/C-N bonds within compound skeletons (Figure 1A). Although methods exist to convert conventional P450 enzymes into P450 peroxidases, successful cases remain scarce and predominantly involve simple reactions. This is primarily attributed to the under-exploration of P450 peroxidase activity and insufficient understanding of its catalytic mechanism. Therefore, in-depth exploration of the catalytic mechanisms of natural P450 peroxidases will facilitate the development of more promising enzymatic reaction systems.
In prior research, the Xu Dong Qu group identified the first P450 enzyme capable of catalysing the dimerisation of tryptophan diketopiperazine (TDKP, Figure 1B) to form pyrroloindole alkaloids (Nat Commun 2018, 9, 4428) and elucidated its unique catalytic mechanism (Nat Commun 2020, 11, 6251). This significantly expanded the substrate spectrum of such P450 enzymes, enabling the biosynthesis of over a hundred bioactive pyrroloindole alkaloids (Angew Chem Int Ed 2023, 62, e202304994), and proposed a novel classification scheme for these alkaloids (Nat Prod Rep, 2022, 39, 1721).
In this study, the research team employed an evolutionarily guided genomic mining strategy to identify a novel P450 enzyme (PtmB) from bacteria. This enzyme catalyses heterodimerisation reactions between three natural purine nucleobases (guanine, hypoxanthine, and adenine) and TDKP, yielding unique C3-nucleobase pyrroloindoles and nucleobase-TDKP dimeric alkaloids. Unlike typical TDKP-dimerising P450 synthases dependent on NAD(P)H cofactors and electron transfer systems, PtmB and its homologues catalyse reactions between adenosine and other 6-amino-containing purine bases with TDKP using H₂O₂, exhibiting rare peroxidase activity.
Through the analysis of multiple PtmB-substrate complex crystal structures, site-directed mutagenesis studies, and QM/MMM calculations, the research team has elucidated the unique catalytic mechanism of PtmB. They discovered that adenine exhibits a dual function in PtmB's peroxidase activity: serving both as the reaction substrate and as an acid-base catalyst that activates H₂O₂ to form the catalytic intermediate Cpd I. Subsequent substrate analogue activity testing, site-directed mutagenesis, and molecular dynamics simulations demonstrated that the NH₂ substituent at the 6-position of adenine is crucial for forming a core base conformation favourable for H₂O₂ activation by participating in an extensive hydrogen-bond network (Figure 1D). Molecular dynamics simulations further revealed flexible conformational transitions in the cyclic dipeptide TDKP, which further promotes the stereoselective and regioselective synthesis of pyrroloindole structures via C-C and C-N bond formation.
This study unveils the first substrate-nucleobase-driven P450 peroxidase catalytic system and represents the most complex chemical reaction known to be catalysed by P450 peroxidases. It enables stereoselective and regioselective construction of C-C and C-N bonds for synthesising intricate natural product skeletons. Nucleobases, fundamental units of DNA and RNA, also function as key acid-base catalysts in certain ribozymes (RNA). This study innovatively uncovers a novel role for nucleobases as acid-base catalysts within P450 peroxidase catalysis. This discovery holds promise for establishing a crucial foundation for developing novel P450 peroxidases and nucleobase-based biocatalysts.
This research was supported by the National Natural Science Foundation of China (NSFC) General Programme, Young Scientist Programme, and Postdoctoral General Fund.
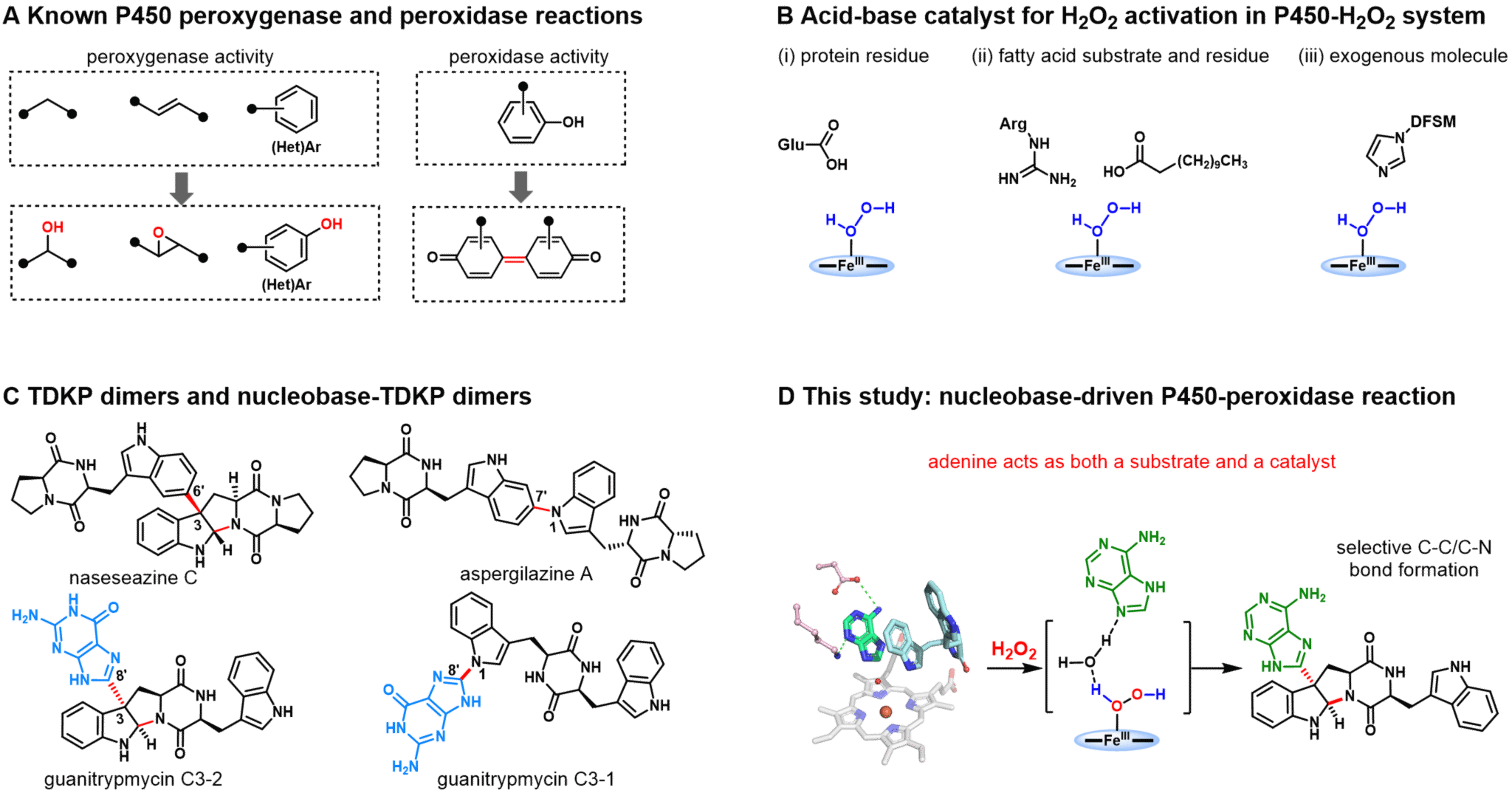
Figure 1. P450-H₂O₂ activity and its mechanism. (A) Comparison of typical P450 peroxygenase and peroxidase activities. (B) Acid-base catalytic mechanisms for H₂O₂ activation in P450 peroxygenase and peroxidase. (C) Representative natural products of TDKP dimers and TDKP nucleobase dimers. (D) Nucleobase-catalysed mechanism identified in this study.
Paper link: https://www.pnas.org/doi/10.1073/pnas.2412890121
Authors: Xu Dong Qu's research group
Contributing unit: Centre for Innovation in Synthetic Science






 Address:No.1308 Keyuan Road, Pudong District, Shanghai
Address:No.1308 Keyuan Road, Pudong District, Shanghai Phone:86-21-54740000
Phone:86-21-54740000 E-mail:zias@sjtu.edu.cn
E-mail:zias@sjtu.edu.cn