搜索


Recently, the research team led by Professor Song Ping from the School of Biomedical Engineering at Shanghai Jiao Tong University and the Zhangjiang Advanced Research Institute (concurrent appointment) published their latest study titled ‘Random Sanitisation in DNA Information Storage Using CRISPR-Cas12a’ in the Journal of the American Chemical Society. This work developed a selective permanent erasure method (RSDISC) combining precise regulation of intelligent nucleic acid probe hybridisation with CRISPR-Cas12a trans-cleavage activity. This technique enables encrypted and protected information storage, validated across nearly 30,000 sequences in multimodal DNA storage—including the Three Character Classic, Tao Te Ching, Art of War, ‘I Have a Dream’ speech, and images of Shanghai Jiao Tong University's Miaomen Gate. Paper link: https://pubs.acs.org/doi/10.1021/jacs.4c11380
With the rapid advancement of the internet, artificial intelligence, and other information technologies, global data volume is projected to reach 175 ZB by 2025. Achieving efficient big data storage has thus become an urgent bottleneck issue. DNA data storage, as an emerging storage technology, is widely regarded as an ideal solution for addressing big data storage demands due to its ultra-high storage density, security, and long-term stability. However, despite its immense potential, DNA storage currently faces numerous challenges in data security. Particularly in big data storage, designing highly secure encryption systems to protect sensitive information and achieving permanent data deletion remain pressing challenges.
Recently, the research group led by Professor Song Ping at Shanghai Jiao Tong University developed a method based on Cas12a trans-cleavage activity and primer-template intelligent nucleic acid selective hybridisation. This approach enables highly sensitive and specific permanent erasure of target information in DNA storage (Figure 1). This approach employs intelligent nucleic acid hybridisation design and regulation to construct reverse primers corresponding to the target file. These primers selectively hybridise and amplify into double strands, thereby protecting the target file. Concurrently, activated Cas12a complexes cleave unprotected single-stranded DNA, enabling precise data erasure. Through further optimisation of nucleic acid hybridisation thermodynamics and Cas12a cleavage activity conditions, researchers validated this method in a multimodal DNA storage system comprising 28,258 oligonucleotides containing images and text. It demonstrated an erasure efficiency of up to 99.9% and specificity of 99.5%. This approach not only safeguards sensitive data but also enables memory cleansing and file classification during big data storage, enhances sequencing accuracy, and holds broad application potential in fields such as molecular diagnostics.
Researchers first validated the method's feasibility in a simple system (Figure 2). Two Cas12a activation systems were designed to assess cleavage efficiency on single templates, with further analysis of their performance on single-stranded DNA exhibiting varying GC content, lengths, and complex secondary structural modifications. Experimental results demonstrated the method's ability to efficiently cleave diverse single-stranded DNA types, achieving up to 99% cleavage efficiency. Subsequently, applying the method to complex multiplex systems featuring template interactions revealed cutting efficiencies exceeding 90% for all templates (Figure 3). Moreover, to evaluate RSDISC's potential for large-scale data cleansing, thermodynamic hybridisation simulations indicated the method could erase up to 15.8 billion files, equivalent to 10 petabytes of data.
To validate the RSDISC method in practical storage systems, the authors further encoded and stored seven files—including the Three Character Classic, The Art of War, the Tao Te Ching, an image of Shanghai Jiao Tong University's gate, and the Mona Lisa—within nearly 30,000 DNA sequences. Experimental results demonstrate that the RSDISC method achieves oligonucleotide erasure efficiency as high as 99.9% with erasure specificity of 99.5% (Figure 4), effectively proving that this method efficiently erases non-target file information while having negligible impact on targeted retained information. This work provides an efficient and reliable information encryption scheme for DNA storage, poised to play a significant role in large-scale data storage and enhancing data processing efficiency in the future.
Shen Hongyu, a doctoral candidate at the School of Biomedical Engineering, Shanghai Jiao Tong University, is the first author of this paper, with Associate Professor Song Ping serving as the corresponding author. This work received funding from the National Key R&D Programme, the National Natural Science Foundation of China, the Central Universities Basic Research Fund, and the Shanghai Municipal Education Commission's ‘Young Leading Talent Cultivation Programme’.
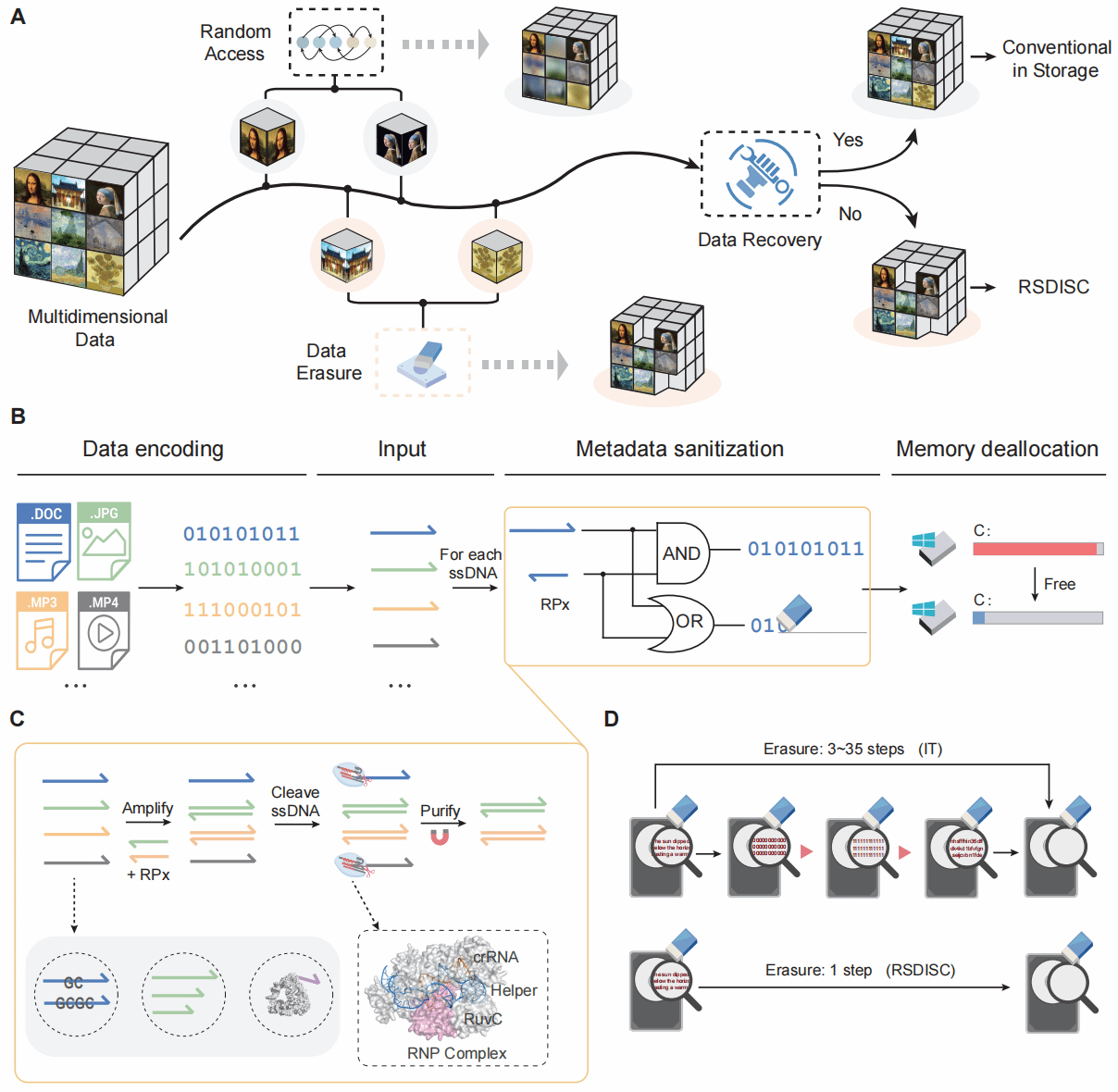
Figure 1: Workflow diagram of the selective erasure method (RSDISC)
Author: Song Ping Research Group
Contributing Unit: DNA Storage Research Centre






 Address:No.1308 Keyuan Road, Pudong District, Shanghai
Address:No.1308 Keyuan Road, Pudong District, Shanghai Phone:86-21-54740000
Phone:86-21-54740000 E-mail:zias@sjtu.edu.cn
E-mail:zias@sjtu.edu.cn